Molecular Analysis of 5-COR Derivatives of Uracil and Evaluating their Affinity Against the MPro Target of COVID-19
DOI:
https://doi.org/10.22034/advjscieng21022079Keywords:
Uracil, Coronavirus, COVID-19, Main protease, Computational, DockingAbstract
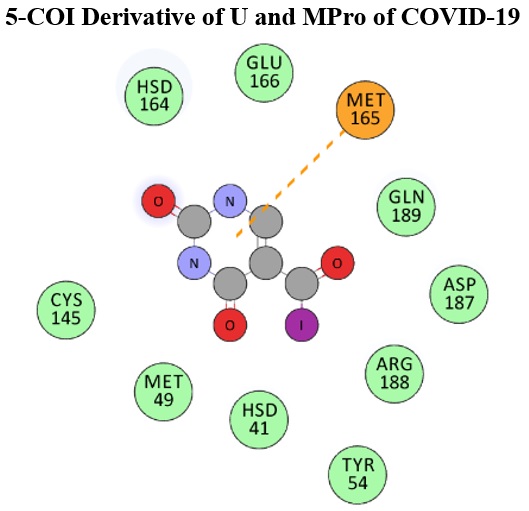
Molecular analysis of 5-COR (R: H, CH3, NH2, OH, F, Cl, Br, I) functionalized derivatives of uracil (U) were explored in this work using computational procedures. Next, binding affinity of each U compound was examined against the main protease (MPro) target of COVID-19 pandemic to maybe inhibit it from further growth. The results indicated that the models of U detected effects of structural modifications by showing variations in their molecular features. Molecular orbital properties indicated that the electronic features of models were changed through functionalization processes. Further analysis of performing molecular docking (MD) simulations also indicated that the models could contribute to different types of interactions with the MPro target, in which the model with 5-COI additional group was highlighted for strong contribution to make the strongest complex of ligand-target system. As a consequence, such structural modification of U helped the models for proper interaction with MPro to maybe inhibit the growth of COVID-19 pandemic.
References
Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737.
Mirzaei M, Panahi MRS, Ghahremani E, Rezae E, Moradi RS, Tahmasebi E, Farnoosh G, Latifi AM. Application of DNA shuffling as a tool for hydrolase activity improvement of pseudomonas strain. Biointerface Research in Applied Chemistry. 2021;11:7735-7745.
Okesola MA, Ogunlana O, Afolabi I, Onasanya A. Ameliorative effect of zingiber officinale on chemical induced DNA damage in rats using PCR analysis. Biointerface Research in Applied Chemistry. 2021;11:11135-11144.
Mirzaei M, Hadipour NL. An investigation of hydrogen-bonding effects on the nitrogen and hydrogen electric field gradient and chemical shielding tensors in the 9-methyladenine real crystalline structure: a density functional theory study. The Journal of Physical Chemistry A. 2006;110:4833-4838.
Yaghoobi R, Mirzaei M. Computational analyses of cytidine and aza-cytidine molecular structures. Lab-in-Silico. 2020;1:21-26.
Behnia S, Nemati F, Fathizadeh S, Salimi M. Heat driven spin currents in DNA chains. Advanced Journal of Science and Engineering. 2020;1:118-121.
Mirzaei M. Effects of carbon nanotubes on properties of the fluorouracil anticancer drug: DFT studies of a CNT-fluorouracil compound. International Journal of Nano Dimension. 2013;3:175-179.
Pyman H, Roshanfekr H, Ansari S. DNA-based electrochemical biosensor using chitosan–carbon nanotubes composite film for biodetection of Pirazon. Eurasian Chemical Communications. 2020;2:213-225.
Mirzaei M, Hadipour NL. Study of hydrogen bonds in crystalline 5-nitrouracil: density functional theory calculations of the O-17, N-14, and H-2 nuclear quadrupole resonance parameters. Journal of the Iranian Chemical Society. 2009;6:195-199.
Roy P. Algorithmic approach to design single strand DNA-based OR logic gate. Lab-in-Silico. 2021;2:44-49.
Abyar Ghamsari P, Samadizadeh M, Mirzaei M. Cytidine derivatives as inhibitors of methyltransferase enzyme. Eurasian Chemical Communications. 2020;2:433-439.
Dhumad AM, Harismah K, Zandi H. Interacting cubane assisted bi-cytidine with COVID-19 main protease: in silico study. Biointerface Research in Applied Chemistry. 2021;11:13962-13967.
Chatterjee P, Lee J, Nip L, Koseki SR, Tysinger E, Sontheimer EJ, Jacobson JM, Jakimo N. A Cas9 with PAM recognition for adenine dinucleotides. Nature Communications. 2020;11:2474.
Mojzeš P, Gao L, Ismagulova T, Pilátová J, Moud?íková Š, Gorelova O, Solovchenko A, Nedbal L, Salih A. Guanine, a high-capacity and rapid-turnover nitrogen reserve in microalgal cells. Proceedings of the National Academy of Sciences. 2020;117:32722-32730.
Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nature Communications. 2020;11:2052.
Li Q, Zhao J, Liu L, Jonchhe S, Rizzuto FJ, Mandal S, He H, Wei S, Sleiman HF, Mao H, Mao C. A poly (thymine)–melamine duplex for the assembly of DNA nanomaterials. Nature Materials. 2020;19:1012-1018.
Felicetti S, Fregoni J, Schnappinger T, Reiter S, de Vivie-Riedle R, Feist J. Photoprotecting uracil by coupling with lossy nanocavities. The Journal of Physical Chemistry Letters. 2020;11:8810-8818.
Harismah K, Mirzaei M, Sahebi H, Gülseren O, Rad AS. Chemically uracil–functionalized carbon and silicon carbide nanotubes: computational studies. Materials Chemistry and Physics. 2018;205:164-170.
Mirzaei M. 5–Fluorouracil: computational studies of tautomers and NMR properties. Turkish Computational and Theoretical Chemistry. 2017;1:27-34.
Mirzaei M, Ahangari RS. Formations of CNT modified 5-(halogen) uracil hybrids: DFT studies. Superlattices and Microstructures. 2014;65:375-379.
Ramesh D, Vijayakumar BG, Kannan T. Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders. European Journal of Medicinal Chemistry. 2020;207:112801.
Ozkendir OM, Askar M, Kocer NE. Influence of the epidemic COVID-19: an outlook on health, business and scientific studies. Lab-in-Silico. 2020;1:26-30.
Harismah K, Mirzaei M. COVID-19: A serious warning for emergency health innovation. Advanced Journal of Science and Engineering. 2020;1:32-33.
Harismah K, Mirzaei M. Favipiravir: structural analysis and activity against COVID-19. Advanced Journal of Chemistry B. 2020;2:55-60.
Dhumad AM, Majeed HJ, Harismah K, Zandi H. In silico approach on ribavirin inhibitors for COVID-19 main protease. Biointerface Research in Applied Chemistry. 2021;11:13924-13933.
Mirzaei M, Harismah K, Da'i M, Salarrezaei E, Roshandel Z. Screening efficacy of available HIV protease inhibitors on COVID-19 protease. Journal Military Medicine. 2020;22:100-107.
Harismaha K, Mirzaeib M, Da'ic M, Roshandeld Z, Salarrezaeid E. In silico investigation of nanocarbon biosensors for diagnosis of COVID-19. Eurasian Chemical Communications. 2021;3:95-102.
Khalid H, Hussain R, Hafeez A. Virtual screening of piperidine based small molecules against COVID-19. Lab-in-Silico. 2020;1:50-55.
Al-Tawfiq JA, Al-Homoud AH, Memish ZA. Remdesivir as a possible therapeutic option for the COVID-19. Travel Medicine and Infectious Disease. 2020;34:101615.
Tobaiqy M, Qashqary M, Al-Dahery S, Mujallad A, Hershan AA, Kamal MA, Helmi N. Therapeutic management of patients with COVID-19: a systematic review. Infection Prevention in Practice. 2020;2:100061.
Mirzaei M. Making sense the ideas in silico. Lab-in-Silico. 2020;1:31-32.
Mirzaei M. Drug discovery: a non-expiring process. Advanced Journal of Chemistry B. 2020;2:46-47.
Soleimanimehr H, Mirzaei M. An introduction to Lab-in-Silico. Lab-in-Silico. 2021;2:1-2.
Kriz K, R?eza?c? J. Benchmarking of semiempirical quantum-mechanical methods on systems relevant to computer-aided drug design. Journal of Chemical Information and Modeling. 2020;60:1453-1460.
Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Montgomery Jr, J.; Vreven, T.; Kudin, K.; Burant, J. Gaussian 09 D.01 Program. Gaussian. Inc.: Wallingford, CT, USA. 2009.
Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289-293.
Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, Feng Z. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research. 2019;47:464-474.
Patil NS, Rohane SH. Organization of Swiss Dock: In study of computational and molecular docking study. Asian Journal of Research in Chemistry. 2021;14:145-148.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Advanced Journal of Science and Engineering

This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License (CC-BY 4.0).



