Molecular Docking of Some Novel Quinoline Derivatives as Potent Inhibitors of Human Breast Cancer Cell Line
DOI:
https://doi.org/10.22034/labinsilico21021030Keywords:
Breast cancer, Binding affinity, Topoisomerase (ii), Doxorubicin, Quinoline, DockingAbstract
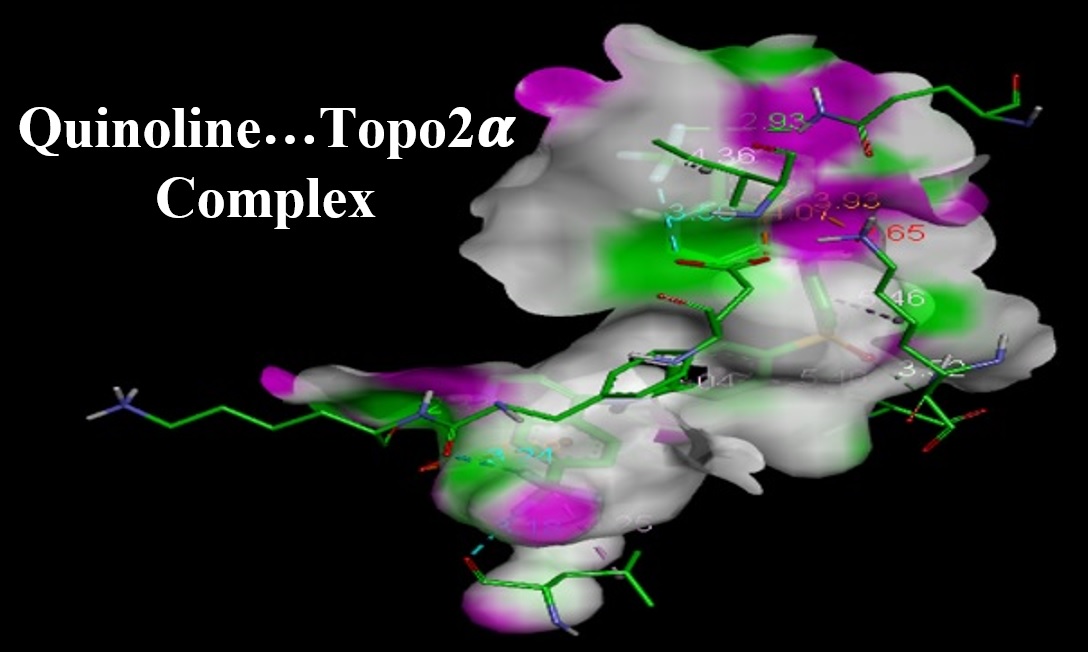
Breast Cancer is one of the major universal health problems affecting more than one million cases per year. Incidence of breast cancer would be seriously increased by inefficacy of the existing available drugs; therefore, designing novel drugs is almost a crucial issue for medication of breast cancer. In this work, some novel synthesized derivatives of quinoline were examined against human breast cancer cell line (MCF-7) through performing structural optimizations and molecular docking simulations to evaluate the binding affinity against topoisomerase (ii) (Topo2????) receptor target. Indeed, first-hand information for the design of novel and potent drugs for medication of breast cancer compounds were provided here. Molecular docking processes were carried out with the help of AutoDock-Vina of PyRx and Discovery Studio software programs. Evaluated binding scores indicated that ligand number 29 could work properly with the lowest binding energy value of -10.4 kcal/mol among 31 investigated ligands. Furthermore, this ligand showed higher binding affinity and bonding strength to the pocket of receptor target (Topo2????) in comparison with the hypothetical Doxorubicin reference drug with binding energy of -6.9 kcal/mol. The provided results of this work could be useful for those researchers working on designing novel medication protocols for breast cancer specially based on quinoline derivatives.
References
Frankish H. 15 million new cancer cases per year by 2020, says WHO. Lancet. 2003;36:1278-1287.
Cancer facts & figures. American Cancer Society. The Society. 2008.
Okobia MN, Bunker CH, Okonofua FE, Osime U. Knowledge, attitude and practice of Nigerian women towards breast cancer: a cross-sectional study. World Journal of Surgical Oncology. 2006;4:11-15.
Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nature Reviews Cancer. 2009;9,327-337.
Pommier Y, Sun Y, Shar-yin NH, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nature Reviews Molecular Cell Biology. 2016;17:703-721.
Deweese JE, Osheroff MA, Osheroff N. DNA topology and topoisomerases: teaching a “Knotty” subject. Biochemistry and Molecular Biology Education. 2009;37:2-10.
Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Research. 2009;37:679-692.
Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of Eukaryotic Topoisomerases in transcription, replication and genomic stability. Nature Reviews Molecular Cell Biology. 2016;17:703-721.
Mukesh B, Rakesh K. Molecular docking: a review. International Journal of Research in Ayurveda and Pharmacy. 2011;2:1746-1751.
Farahbakhsh Z, Zamani MR, Rafienia M, Gülseren O, Mirzaei M. In silico activity of AS1411 aptamer against nucleolin of cancer cells. Iranian Journal of Blood and Cancer. 2020;12:95-100.
Guedes IA, de Magalhães CS, Dardenne LE. Receptor-ligand molecular docking. Biophysical Reviews. 2014;6:75-87.
Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prli? A, Quesada M, Quinn GB, Westbrook JD, Young J. The RCSB protein data bank: redesigned web site and web services. Nucleic Acids Research. 2010;39:392-401.
Mirali M, Jafariazar Z, Mirzaei M. Loading tacrine Alzheimer’s drug at the carbon nanotube: DFT approach. Lab-in-Silico. 2021 3;2:3-8.
Khalid H, Hussain R, Hafeez A. Virtual screening of piperidine based small molecules against COVID-19. Lab-in-Silico. 2020;1:50-55.
Mirzaei M, Harismah K, Da’I M, Salarrezaei E, Roshandel Z. Screening efficacy of available HIV protease inhibitors on COVID-19 protease. Journal Military Medicine. 2020;22:100-107.
Harismah K, Mirzaei M. Steviol and iso-steviol vs. cyclooxygenase enzymes: in silico approach. Lab-in-Silico. 2020;6:11-15.
Dizdaroglu Y, Albay C, Arslan T, Ece A, Turkoglu EA, Efe A, Senturk M, Supuran CT, Ekinci D. Design, synthesis and molecular modelling studies of some pyrazole derivatives as carbonic anhydrase inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry. 2020;35:289-297.
Yamali C, Gul HI, Ece A, Bua S, Angeli A, Sakagami H, Sahin E, Supuran CT. Synthesis, biological evaluation and in silico modelling studies of 1, 3, 5-trisubstituted pyrazoles carrying benzenesulfonamide as potential anticancer agents and selective cancer-associated hCA IX isoenzyme inhibitors. Bioorganic Chemistry. 2019;92:103222.
Mirzaei M, Hadipour NL. An investigation of hydrogen-bonding effects on the nitrogen and hydrogen electric field gradient and chemical shielding tensors in the 9-methyladenine real crystalline structure: a density functional theory study. The Journal of Physical Chemistry A. 2006;110:4833-4838.
Harismah K, Ozkendir OM, Mirzaei M. Lithium adsorption at the C20 fullerene-like cage: DFT approach. Advanced Journal of Science and Engineering. 2020;1:74-79.
Mirzaei M. Science and engineering in silico. Advanced Journal of Science and Engineering. 2020;1:1-2.
Mirzaei M. Making sense the ideas in silico. Lab-in-Silico. 2020;1:31-32.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Lab-in-Silico

This work is licensed under a Creative Commons Attribution 4.0 International License.




