Hydrogen Bond Interactions of Nucleobases: A Quick Review
DOI:
https://doi.org/10.22034/labinsilico20012061Keywords:
Hydrogen bond, Interaction, Nucleobase, DFT, DNA, RNAAbstract
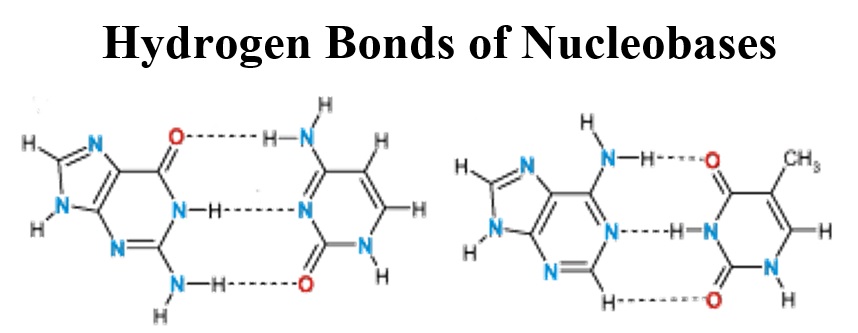
Hydrogen bond (HB) interactions in nucleobases; Adenine (A), Guanine (G), Cytosine (C), Thymine (T), and Uracil (U), were discussed quickly in this review work. The features of HB interactions were already investigated by the means of computer-based works and experiments. Formations of homo and hetero complexes of nucleobases through HB interactions could be characterizations using obtained parameters from X-ray, NMR and NQR techniques. Such spectroscopic properties could be evaluated by both of computations and experiments. Indeed, structural refinement could be also done by computations, in which more reliable results could be provided for further investigations. In addition to original nucleobases, their modified structures could also work their roles especially in medications. Due to such importance of the topic, the features of HB interactions in nucleobases were discussed in this quick review.
References
Watson JD, Crick FH. A structure for deoxyribose nucleic acid. Nature. 1953;171:737-738.
Kazazian Jr HH, Moran JV. Mobile DNA in health and disease. New Engl J Med. 2017;377:361-370.
Li W. The study of correlation structures of DNA sequences: a critical review. Comp Chem. 1997;21:257-271.
Cao C, Yu J, Li MY, Wang YQ, Tian H, Long YT. Direct readout of single nucleobase variations in an oligonucleotide. Small. 2017;13:1702011.
Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2005;7: 61-77.
Mirzaei M, Yousefi M. Computational studies of the purine-functionalized graphene sheets. Superlatt Microstruct. 2012;52:612-617.
Mirzaei M, Hadipour NL. Study of hydrogen bonds in crystalline 5-nitrouracil. Density functional theory calculations of the O-17, N-14, and H-2 nuclear quadrupole resonance parameters. J Iran Chem Soc. 2009;6:195-199.
Fersht AR. The hydrogen bond in molecular recognition. Trends Biochem Sci. 1987;12:301-304.
Samadi Z, Mirzaei M, Hadipour NL, Khorami SA. Density functional calculations of oxygen, nitrogen and hydrogen electric field gradient and chemical shielding tensors to study hydrogen bonding properties of peptide group (OC–NH) in crystalline acetamide. J Mol Graph Model. 2008;26:977-981.
Rofouei MK, Soleymani R, Aghaei A, Mirzaei M. Synthesis, vibrational, electrostatic potential and NMR studies of (E and Z) 1-(4-chloro-3-nitrophenyl)-3-(2-methoxyphenyl) triazene: Combined experimental and DFT approaches. J Mol Struct. 2016;1125:247-259.
Gilani AG, Taghvaei V, Rufchahi EM, Mirzaei M. Tautomerism, solvatochromism, preferential solvation, and density functional study of some heteroarylazo dyes. J Mol Liquid. 2019;273:392-407.
Šponer J, Hobza P. Molecular interactions of nucleic acid bases. A review of quantum-chemical studies. Collect Czech Chem Commun. 2003;68:2231-2282.
Šponer J, Leszczynski J, Hobza P. Hydrogen bonding and stacking of DNA bases: a review of quantum-chemical ab initio studies. J Biomol Struct Dyn. 1996;14:117-135.
Aghazadeh M, Mirzaei M. Hydrogen bond interactions in sulfamerazine: DFT study of the O-17, N-14, and H-2 electric field gradient tensors. Chem Phys. 2008;351:159-162.
Savelev I, Myakishev-Rempel M. Evidence for DNA resonance signaling via longitudinal hydrogen bonds. Prog Biophys Mol Biol. 2020;156:14-19.
Sarkar S, Singh PC. Alteration of the groove width of DNA induced by the multimodal hydrogen bonding of denaturants with DNA bases in its grooves affects their stability. Biochim Biophys Acta. 2020;1864:129498.
Gordon MS, Jensen JH. Understanding the hydrogen bond using quantum chemistry. Acc Chem Res. 1996;29:536-543.
Ireta J, Neugebauer J, Scheffler M. On the accuracy of DFT for describing hydrogen bonds: dependence on the bond directionality. J Phys Chem A. 2004;108:5692-5698.
Mirzaei M, Gülseren O, Hadipour N. DFT explorations of quadrupole coupling constants for planar 5-fluorouracil pairs. Comput Theor Chem. 2016;1090:67-73.
Mirzaei M, Kalhor HR, Hadipour NL. Covalent hybridization of CNT by thymine and uracil: A computational study. J Mol Model. 2011;17:695-699.
Mirzaei M, Hadipour NL. An investigation of hydrogen-bonding effects on the nitrogen and hydrogen electric field gradient and chemical shielding tensors in the 9-methyladenine real crystalline structure: a density functional theory study. J Phys Chem A. 2006;110:4833-4838.
van Mourik T, Dingley AJ. Characterization of the monovalent ion position and hydrogen?bond network in guanine quartets by DFT calculations of NMR parameters. Chem Eur J. 2005;11:6064-6079.
Mirzaei M, Elmi F, Hadipour NL. A systematic investigation of hydrogen-bonding effects on the 17O, 14N, and 2H nuclear quadrupole resonance parameters of anhydrous and monohydrated cytosine crystalline structures: a density functional theory study. J Phys Chem B. 2006;110:10991-10996.
Mirzaei M, Hadipour NL. A computational NQR study on the hydrogen?bonded lattice of cytosine?5?acetic acid. J Comput Chem. 2008;29:832-838.
Mirzaei M, Hadipour NL, Ahmadi K. Investigation of C–H… OC and N–H… OC hydrogen-bonding interactions in crystalline thymine by DFT calculations of O-17, N-14 and H-2 NQR parameters. Biophys Chem. 2007;125:411-415.
Mirzaei M, Hadipour NL. Study of hydrogen bonds in 1-methyluracil by DFT calculations of oxygen, nitrogen, and hydrogen quadrupole coupling constants and isotropic chemical shifts. Chem Phys Lett. 2007;438:304-307.
Johnston RC, Cheong PH. C–H?O non-classical hydrogen bonding in the stereomechanics of organic transformations: theory and recognition. Org Biomol Chem. 2013;11:5057-5064.
Partovi T, Mirzaei M, Hadipour NL. The C–H?O hydrogen bonding effects on the 17O electric field gradient and chemical shielding tensors in crystalline 1-methyluracil: A DFT study. Z Naturforsch A. 2006;61:383-388.
Steiner T. The hydrogen bond in the solid state. Angew Chem. 2002;41:48-76.
Bagno A, Saielli G. Computational NMR spectroscopy: reversing the information flow. Theor Chem Acc. 2007;117:603-619.
Mirzaei M. The NMR parameters of the SiC-doped BN nanotubes: a DFT study. Physica E. 2010;42:1954-1957.
Mirzaei M, Hadipour NL, Abolhassani MR. Influence of C-doping on the B-11 and N-14 quadrupole coupling constants in boron-nitride nanotubes: A DFT study. Z Naturforsch A. 2007;62:56-60.
Bodaghi A, Mirzaei M, Seif A, Giahi M. A computational NMR study on zigzag aluminum nitride nanotubes. Physica E. 2008;41:209-212.
Seif A, Mirzaei M, Aghaie M, Boshra A. AlN nanotubes: A DFT study of Al-27 and N-14 electric field gradient tensors. Z Naturforsch A. 2007;62:711-715.
Mirzaei M, Nouri A, Giahi M, Meskinfam M. Computational NQR study of a boron nitride nanocone. Monatsh Chem. 2010;141:305-307.
Harismah K, Mirzaei M, Sahebi H, Gülseren O, Rad AS. Chemically uracil–functionalized carbon and silicon carbide nanotubes: Computational studies. Mater Chem Phys. 2018;205:164-170.
Mirzaei M, Kalhor HR, Hadipour NL. Investigating purine-functionalised carbon nanotubes and their properties: a computational approach. IET Nanobiotechnol. 2011;5:32-35.
Faramarzi R, Falahati M, Mirzaei M. Interactions of fluorouracil by CNT and BNNT: DFT analyses. Adv J Sci Eng. 2020;1:62-66.
Mirzaei M, Ravi S, Yousefi M. Modifying a graphene layer by a thymine or a uracil nucleobase: DFT studies. Superlatt Microstruct. 2012;52:306-311.
Mirzaei M. Uracil-functionalized ultra-small (n, 0) boron nitride nanotubes (n= 3–6): Computational studies. Superlatt Microstruct. 2013;57:44-50.
Ghanadian M, Ali Z, Khan IA, Balachandran P, Nikahd M, Aghaei M, Mirzaei M, Sajjadi SE. A new sesquiterpenoid from the shoots of Iranian Daphne mucronata Royle with selective inhibition of STAT3 and Smad3/4 cancer-related signaling pathways. DARU. 2020;28:253-262.
Fonseca Guerra C, Bickelhaupt FM, Snijders JG, Baerends EJ. Hydrogen bonding in DNA base pairs: reconciliation of theory and experiment. J Am Chem Soc. 2000;122:4117-4128.
Petrovykh DY, Kimura-Suda H, Tarlov MJ, Whitman LJ. Quantitative characterization of DNA films by X-ray photoelectron spectroscopy. Langmuir. 2004;20:429-440.
Harismah K, Ozkendir OM, Mirzaei M. Explorations of crystalline effects on 4-(benzyloxy)benzaldehyde properties. Z Naturforsch A. 2015;70:1013-1018.
Ida R, De Clerk M, Wu G. Influence of N–H?O and C–H?O hydrogen bonds on the 17O NMR tensors in crystalline uracil: Computational study. J Phys Chem A. 2006;110:1065-1071.
Wu G, Dong S, Ida R. Solid-state 17O NMR of thymine: a potential new probe to nucleic acid base pairing. Chem Commun. 2001:891-892.
Dra?ínský M, Hodgkinson P. Solid-state NMR studies of nucleic acid components. RSC Adv. 2015;5:12300-12310.
Wu G, Dong S, Ida R, Reen N. A solid-state 17O nuclear magnetic resonance study of nucleic acid bases. J Am Chem Soc. 2002;124:1768-1777.
Martic S, Wu G, Wang S. Interactions of cytidine with N2-functionalized guanosines and cytidine–cytidine exchange involving a GC pair—NMR and fluorescence spectroscopic study. Canadian J Chem. 2010;88:524-532.
Ida R, Wu G. Direct NMR detection of alkali metal ions bound to G-quadruplex DNA. J Am Chem Soc. 2008;130:3590-3602.
Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900-1909.
Harismah K, Mirzaei M. In silico interactions of steviol with monoamine oxidase enzymes. Lab-in-Silico. 2020;1:3-6.
Harismah K, Mirzaei M. Steviol and iso-steviol vs. cyclooxygenase enzymes: In silico approach. Lab-in-Silico. 2020;1:11-15.
Khalid H, Hussain R, Hafeez A. Virtual screening of Piperidine based small molecules against COVID-19. Lab-in-Silico 2020;1:50-55.
Yaghoobi R, Mirzaei M. Computational analyses of cytidine and aza-cytidine molecular structures. Lab-in-Silico. 2020;1:21-25.
Mirzaei M, Harismah K, Da'i M, Salarrezaei E, Roshandel Z. Screening efficacy of available HIV protease inhibitors on COVID-19 protease. J Mil Med. 2020;22:100-107.
Farahbakhsh Z, Zamani MR, Rafienia M, Gulseren O, Mirzaei M. In silico activity of AS1411 aptamer against nucleolin of cancer cells. Iran J Blood Cancer. 2020;12:95-100.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2020 Lab-in-Silico

This work is licensed under a Creative Commons Attribution 4.0 International License.




